Proof of concept for querying spatial data on IPFS using geohash#
InterPlanetary File System (IPFS) offers an alternative approach to store and distribute spatial data. Size of spatial data grows exponentially so it is imperative to perform efficient partial selection of data to avoid large bulk download over the IPFS which can save both transaction time and local storage space. Yet there is a lack of support in spatial query functionality like PostGIS in the traditional database of PostgreSQL. This blog post introduces one type of spatial indexing – geohash and walks through a proof of concept to integrate geohash into the IPFS system. And the final section depicts a preliminary result for the implementation in comparison with solutions using local storage, local database, and vanilla IPFS without any spatial index.
Geohash#
Geohash is a public domain geocode system invented in 2008 by Gustavo Niemeyer which encodes a geographic location into a short string of letters and digits. Similar ideas were introduced by G.M. Morton in 1966. It is a hierarchical spatial data structure which subdivides space into buckets of grid shape, which is one of the many applications of what is known as a Z-order curve, and generally space-filling curves.
Compared to other prevalent hierarchical spatial data structure like R-tree and quadtree, geohash is using an absolute coordinate-encoding mapping which does not change value by the extent of dataset.
Following code snippets show basic usage of the Python package pygeohash to encode a coordinates to geohash and to decode a geohash to the centeriod coordinates
[ ]:
#!pip install pygeohash (https://github.com/wdm0006/pygeohash)
import pygeohash as pgh
pgh.encode(latitude=42.6, longitude=-5.6)
# >>> 'ezs42e44yx96'
pgh.encode(latitude=42.6, longitude=-5.6, precision=5)
# >>> 'ezs42'
pgh.decode(geohash='ezs42')
# >>> ('42.6', '-5.6')
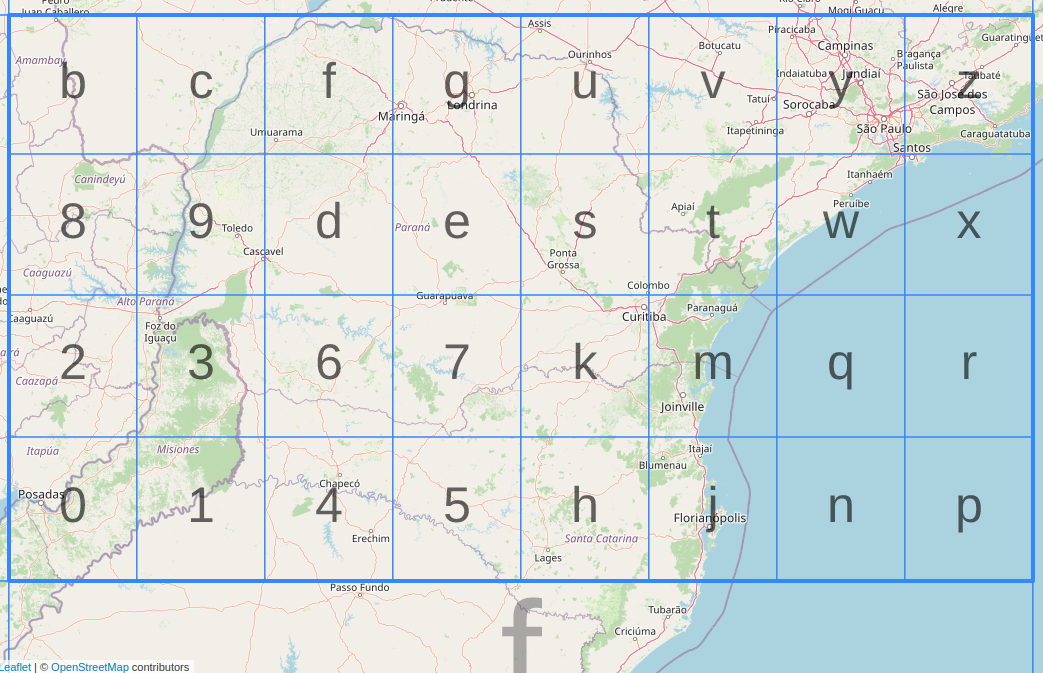
Proof of concept: Mapping geohash with spatial asset via index folder#
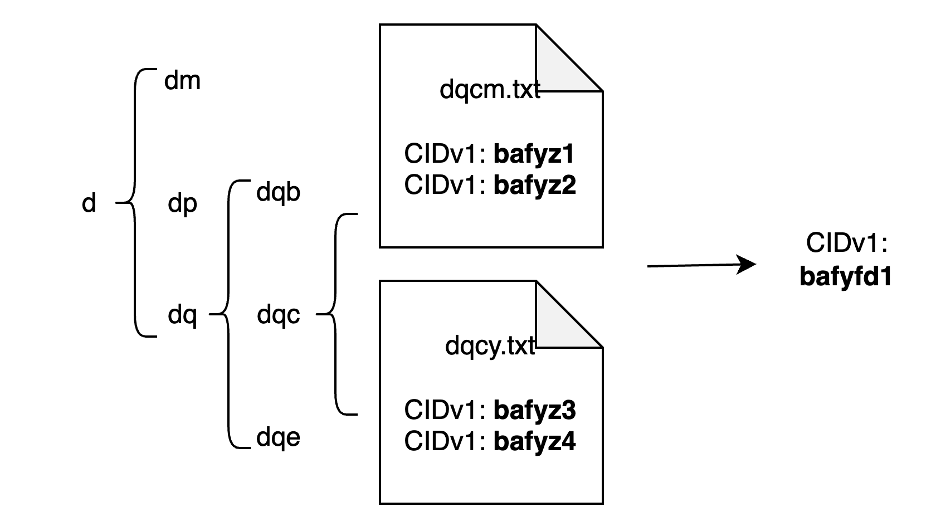
The most straightforward solution is to create a hierarchical folder structure with subfolder names representing the geohash subgrids within the parent grid. The leaf node folder refers to geohash with the finest resolution, and contains CIDs that can point to the asset located in the leaf geohash. The entire folder would be packed and uploaded to IPFS as a separate asset. The query process consists of two steps: locate the target geohash; and recursively collect all the CIDs under the geohash node. Using the CIDs from the query steps, a partial retrieval can be achieved. Here is an example of 2600 restaurant points in Washington, DC, downloaded from OpenStreetMap.
[ ]:
##load geojson
import geopandas as gpd
import pandas as pd
import os
load point data and assign geohash to each point#
[ ]:
#demo_points = gpd.read_file(f"../../data/maryland_demo/dc_restaurants.geojson")
demo_points = pd.concat([demo_points,demo_points.get_coordinates()],axis=1)
demo_points['geohash'] = demo_points.apply(lambda row: pgh.encode(row['y'], row['x'],precision=6), axis=1)
“shatter” the points as individual geojson and calculate CIDs#
NOTE: make sure IPFS daemon is running before calculating CIDs
[ ]:
# Directory where the individual GeoJSON files will be saved
asset = "dc_restaurants"
directory = f"../data/geohash_{asset}"
# Make sure the directory exists, if not create it
if not os.path.exists(directory):
os.makedirs(directory)
# Initialize an empty list to store file paths
file_paths = []
# Loop through each row in GeoDataFrame
for index, row in demo_points.iterrows():
# Slice the GeoDataFrame to get a single feature (row)
single_feature_gdf = demo_points.iloc[[index]]
# Get 'osm_id' for the single feature
osm_id = row['osm_id']
# Define the full file path
file_path = os.path.join(directory, f"{osm_id}.geojson")
# Save single feature GeoDataFrame as GeoJSON
single_feature_gdf.to_file(file_path, driver="GeoJSON")
# Append file_path to list
file_paths.append(file_path)
# Create a new column in the original GeoDataFrame to store file paths
demo_points['single_path'] = file_paths
[ ]:
#make sure ipfs daemon is running!!
def compute_cid(file_path):
import subprocess
cid = subprocess.check_output(["ipfs", "add", "-qn", file_path]).decode().strip()
return cid
demo_points['single_cid'] = demo_points.apply(lambda x: compute_cid(x['single_path']),axis=1)
create index folder with a trie data structure#
[ ]:
class TrieNode:
def __init__(self):
self.children = {}
self.value = []
class Trie:
def __init__(self):
self.root = TrieNode()
def insert(self, index, value):
node = self.root
for char in str(index):
if char not in node.children:
node.children[char] = TrieNode()
node = node.children[char]
node.value.append(value)
def get(self, index):
node = self.root
for char in str(index):
if char not in node.children:
return None
node = node.children[char]
return node.value
[ ]:
#arrange cid by geohash
pairs = list(zip(demo_points['geohash'],demo_points['single_cid']))
# Create an empty Trie dictionary
trie_dict = Trie()
# Insert each index-value pair into the Trie dictionary
for index, value in pairs:
trie_dict.insert(index, value)
[ ]:
def export_trie(trie_node,geohash,root_path):
#export geojson at current hash level
next_path = root_path+"/"+"".join(geohash)
leaf_path = root_path+f"/{geohash}.txt"
print(geohash,root_path,next_path,leaf_path)
if trie_node.value:
# Open a file in write mode
with open(leaf_path, 'w') as f:
for item in trie_node.value:
f.write(f"{item}\n")
#make path and export to sub folder
import os
if trie_node.children and not os.path.exists(next_path):
os.makedirs(next_path)
for ch in trie_node.children:
child_hash = geohash+ch
export_trie(trie_node.children[ch],child_hash,next_path)
export_trie(trie_dict.root,"",f"../data/geohash_{asset}/index")
Performance benchmarking: query queen neighbors from a given geohash#
Example query to retrieve 329 out of 2675 point features: the target is to get nearby restaurants (green dots) from the neighboring geohash (blue grids) of a selected standing point (red dot) within a geohash grid (dqcjy)
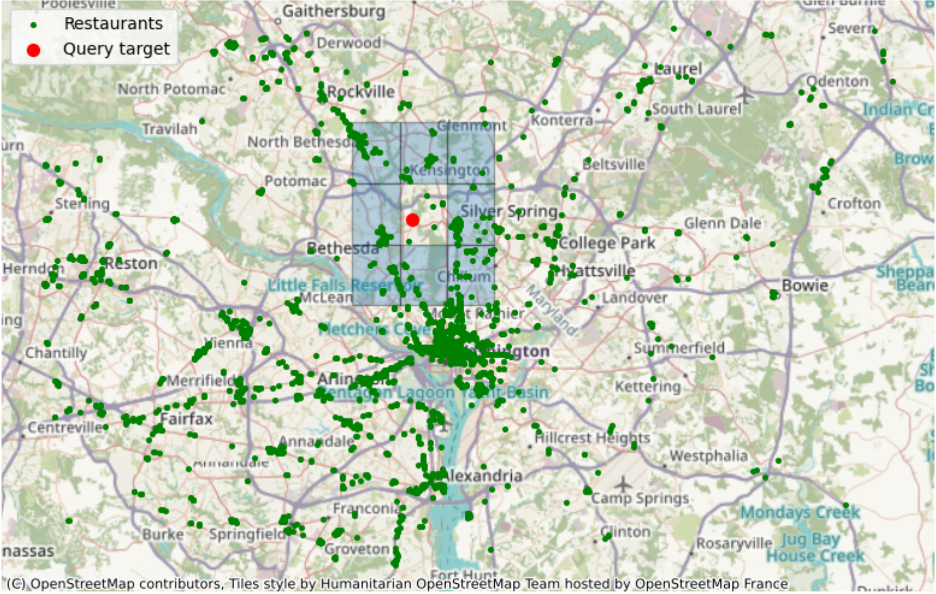
Calculate queen neighbor#
[ ]:
def queen_neighbors(geohash: str) -> list:
import pygeohash as pgh
nei = rook_neighbors(geohash)
directions = ["right","bottom","left","top"]
for i in range(4):
nei.append(pgh.get_adjacent(nei[i],directions[i]))
return nei
q_geohash = "dqcjy"
qn = queen_neighbors(q_geohash)
Query the cid list within the queen neighbor geohashes#
[3]:
# load index structure
## from file structure
def compose_path(s,root):
"""
compose path a/ab/abc for geohash `abc`
"""
path = [root]
for i in range(len(s)):
path.append(s[:i+1])
return "/".join(path)
def process_leaf_node(leaf):
"""
process index leaf.
leaf: txt file path of a index leaf, like a//ab/abc.txt
"""
with open(leaf, 'r', encoding='utf-8') as file:
lines = file.readlines()
return [line.strip() for line in lines]
def traverse_sub_node(node):
"""
recursively collect all the leaf node under the current node
"""
import os
results=[]
excludes = [".ipynb_checkpoints"]
# Get list of items in the directory
subfolders = [d for d in os.listdir(node) if os.path.isdir(os.path.join(node, d)) and d not in excludes]
# If there are subfolders, traverse them
if subfolders:
for subfolder in subfolders:
results.extend(traverse_sub_node(os.path.join(node, subfolder)))
else:
# Otherwise, process txt files in the directory
txt_files = [f for f in os.listdir(node) if f.endswith('.txt')]
for txt_file in txt_files:
results.extend(process_leaf_node(os.path.join(node, txt_file)))
return results
def query_feature_cid_by_geohash(geohash: str, index_root: str) -> list:
"""
find matching geohash or sub-level hashs
"""
import os
target_path = compose_path(geohash,index_root)
cid_list = []
if os.path.exists(target_path):
cid_list = traverse_sub_node(target_path)
if os.path.exists(target_path+'.txt'):
cid_list = process_leaf_node(target_path+'.txt')
return cid_list
def ipfs_get_feature(cid):
import subprocess
subprocess.check_output(["ipfs", "get", cid])
return gpd.read_file(f"./{cid}")
Benchmark for ipfs+geohash#
[ ]:
%%timeit
import subprocess
subprocess.check_output(["ipfs", "get", "-o","../data/test/dc","QmPaXWva3WQR2uwFdu6bkizUyRyKpX3V1aiT5dvnnYKSpJ"])
#query
results = multi_geohash_query(rn,f"../data/test/dc")
os.chdir("../data/test/")
#ipfs retrieval
ipfs_retrieval = pd.concat([ipfs_get_feature(cid) for cid in results])
os.chdir("../../notebooks")
Benchmark for local I/O+geohash#
[ ]:
%%timeit
#query
results = multi_geohash_query(rn,f"../data/geohash_{asset}/index")
local_io_retrieval = pd.concat([gpd.read_file(path) for path in gdf[gdf.single_cid.isin(results)].single_path.tolist()])
Benchmark for geopandas query#
[ ]:
%%timeit
#candidate_points = gpd.read_file(f"../../data/maryland_demo/{asset}_cid.geojson")
gpd.sjoin(candidate_points, neighbors, how="inner", op="within")
Benchmark for postgresql#
run docker daemon and the command to spin up postgresql docker-compose up
[ ]:
import geopandas as gpd
from sqlalchemy import create_engine
import psycopg2
# Load GeoJSON into a GeoDataFrame
gdf = gpd.read_file(f"../../data/maryland_demo/{asset}.geojson")
# Connect to PostgreSQL
engine = create_engine('postgresql://user:password@localhost:5432/geodb')
# Load data into PostgreSQL
gdf.to_postgis(f'{asset}', engine, if_exists='replace', index=False)
[ ]:
##Create spatial index
# Parameters for connection
params = {
'dbname': 'geodb',
'user': 'user',
'password': 'password',
'host': 'localhost',
'port': '5432'
}
# Create a connection and cursor
conn = psycopg2.connect(**params)
cur = conn.cursor()
# Execute the CREATE INDEX command
cur.execute(f'CREATE INDEX ON {asset} USING gist(geometry)')
# Commit the changes and close the connection
conn.commit()
cur.close()
conn.close()
[ ]:
# store query target geodataframe into database
neighbors.crs = "EPSG:4326"
neighbors.to_postgis("temp_table", engine, if_exists="replace")
[ ]:
#benchmark
%%timeit
conn = psycopg2.connect(**params)
# Create a new cursor
cur = conn.cursor()
# Execute a COUNT SQL
sql = f"""
SELECT COUNT(*) FROM
(SELECT {asset}.geometry
FROM {asset}, temp_table
WHERE ST_Intersects({asset}.geometry, temp_table.geometry)) AS R;
"""
cur.execute(sql)
# Fetch the result
count = cur.fetchone()[0]
# Close the cursor and connection
cur.close()
conn.close()
Benchmark comparison#
Query Method for queen neighbor proximity |
Number of nearby features (per loop time of 7 runs, 10 loops each) |
Retrieval of nearby features |
|---|---|---|
Geohash + local I/O |
0.77 ms |
3190 ms |
Geohash + IPFS |
257 ms |
13500 ms |
In-memory query (geopandas.sjoin ) |
20 ms |
14600 ms |
Postgis |
21.3 ms |
18.8 ms |
From the results of local I/O, we can see pre-calculated geohash can significant reduce the query time at the specific task. However, IPFS online test indicates the I/O expense is still under optimized for IPFS system when retrieving the index file is inevitable. When including the file retrieval, partial retrieval with geohash overperformed the in-memory query where geohash is not available and all the assets need to be loaded into the memory. Yet local database system is still the optimized option when considering the retrieval process implying improvment in file exchange process within IPFS.
Try yourself#
To replicate the code locally, you may check out the project repository for environment setup.
